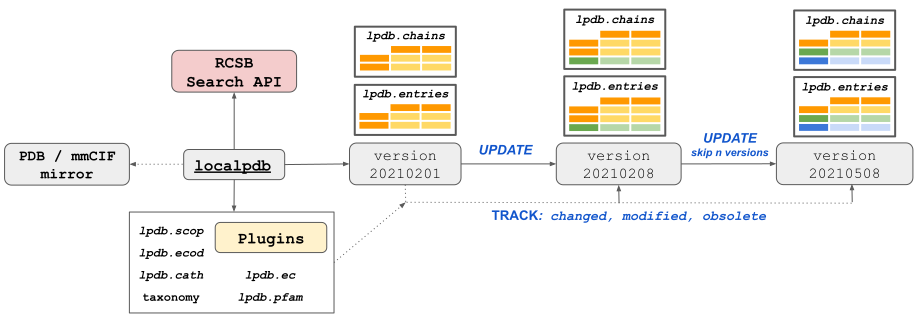
localpdb
localpdb provides a simple framework to store the local mirror of the protein structures available in the PDB database and other related resources.
The underlying data can be conveniently browsed and queried with the pandas.DataFrame structures.
Update mechanism allows following the weekly PDB releases while retaining the possibility to access previous data versions.
You may find localpdb particularly useful if you:
- already use Biopython
Bio.PDB.PDBListor similar modules and tools like CCPDB, - build custom protein datasets based on multiple criteria, e.g. for machine learning purposes,
- create pipelines based on the multiple or all available protein structures,
- are a fan of pandas
DataFrames.
Overview
 To find more about the package and its functionalities please follow the docs. In case of any troubles free to contact us or open an issue.
To find more about the package and its functionalities please follow the docs. In case of any troubles free to contact us or open an issue.
Installation
pip install localpdb
localpdb_setup -db_path /path/to/localpdb --fetch_cif
Examples
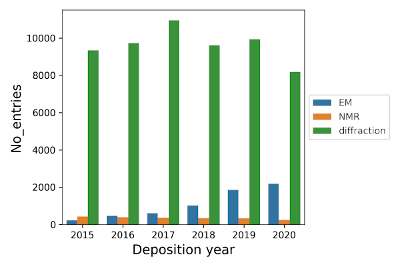
Find the number of entries added to the PDB every year
from localpdb import PDB
lpdb = PDB(db_path='/path/to/your/localpdb')
lpdb.entries = lpdb.entries.query('deposition_date.dt.year >= 2015 & deposition_date.dt.year <= 2020')
df = lpdb.entries.groupby(by=['method', lpdb.entries.deposition_date.dt.year])['mmCIF_fn'].count().reset_index()
sns.barplot(data=df, x='deposition_date', y='mmCIF_fn', hue='method')
Create a custom dataset of protein chains
Select:
- human SAM-dependent methyltransferases,
- solved with X-ray diffraction,
- with resolution below 2.5 Angstrom
- deposited after 2010.
- remove the redundancy at the 90% sequence identity,
# Install plugins providing additional data localpdb_setup -db_path /path/to/your/localpdb -plugins SIFTS ECOD PDBClusteringfrom localpdb import PDB import gzip lpdb.entries = lpdb.entries.query('type == "prot"') # Protein structures lpdb.entries = lpdb.entries.query('method == "diffraction"') # solved with X-ray diffraction lpdb.entries = lpdb.entries.query('resolution <= 2.5') # with resolution below 2.5A lpdb.entries = lpdb.entries.query('deposition_date.dt.year >= 2010') # added after 2010 lpdb.chains = lpdb.chains.query('ncbi_taxid == "9606"') # human proteins lpdb.ecod = lpdb.ecod.query('t_name == "S-adenosyl-L-methionine-dependent methyltransferases"') # SAM dependent methyltransferases # Remove redundancy (select only representative structure from each sequence cluster) lpdb.load_clustering_data(redundancy=90) lpdb.chains = lpdb.chains[lpdb.chains['clust-90'].notnull()] representative = lpdb.chains.groupby(by='clust-90')['resolution'].idxmin() lpdb.chains = lpdb.chains.loc[representative] lpdb.chains.to_csv('dataset.csv') # Save dataset
Advanced examples
-
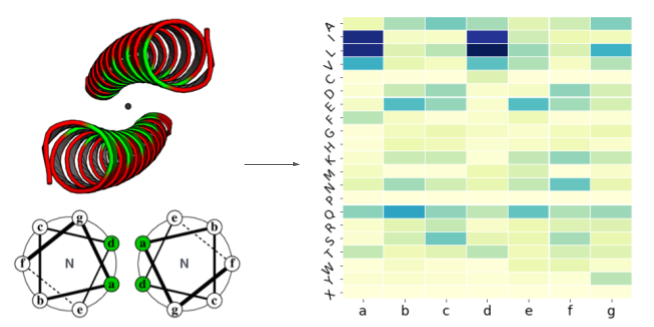
Dynamics of the HIV protease inferred from the PDB structures.
-
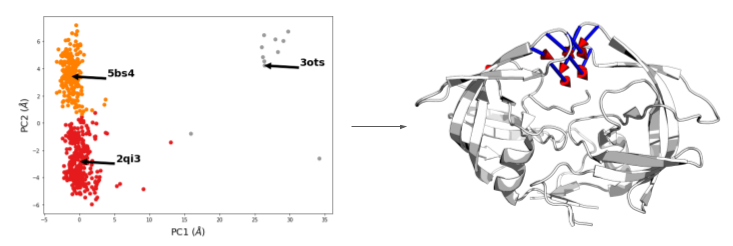
Creating a dataset for machine learning - example of building DeepCoil training and test sets.
Acknowledgments
This work was supported by the National Science Centre grant 2017/27/N/NZ1/00716.